read_ref_file="sacB_ref.fasta"
ref=next(SeqIO.parse(os.path.join(data_path,read_ref_file),"fasta"))
#read_ref_file="\\sacB_ref.fasta"
#ref=next(SeqIO.parse(data_path+read_ref_file,'fasta'))
ref_seq=str(ref.seq)
gen_list=parse_genotypes(os.path.join(data_path,"sacB_genotypes.csv"))plotting
Functions to plot mutation results
# read_ref_file_paul='/home/prochett/DGRec/dgrec/example_data/sacB_ref.fasta'
# ref_paul=next(SeqIO.parse(os.path.join(data_path,read_ref_file_paul),"fasta"))
# ref_seq=str(ref_paul.seq)
# gen_list=[]
# with open(os.path.join(data_path,"/home/prochett/DGRec/dgrec/example_data/sacB_genotypes.csv"),"r") as handle:
# reader = csv.reader(handle, delimiter='\t')
# for row in reader:
# gen_list.append((row[0],int(row[1])))plot_mutations
plot_mutations (gen_list:list, ref_seq:str, sample_name:str=None, plot_range:Union[tuple,list]=None, TR_range:Union[tuple,list]=None, ax=None)
| Type | Default | Details | |
|---|---|---|---|
| gen_list | list | list of genotypes. Each genotype is a tuple: (string representation of the genotype, number of molecules) | |
| ref_seq | str | reference sequence | |
| sample_name | str | None | sample name |
| plot_range | Union | None | limits the plot to the specified range |
| TR_range | Union | None | when specified creates a shaded box highlighting the position of the TR |
| ax | NoneType | None | makes it possible to pass matplotlib axis to easily configure and save plots |
plot_mutations_percentage
plot_mutations_percentage (gen_list:list, ref_seq:str, sample_name:str=None, plot_range:Union[tuple,list]=None, TR_range:Union[tuple,list]=None, rev_comp=False, ax=None)
| Type | Default | Details | |
|---|---|---|---|
| gen_list | list | list of genotypes. Each genotype is a tuple: (string representation of the genotype, number of molecules) | |
| ref_seq | str | reference sequence | |
| sample_name | str | None | sample name |
| plot_range | Union | None | limits the plot to the specified range |
| TR_range | Union | None | when specified creates a shaded box highlighting the position of the TR |
| rev_comp | bool | False | |
| ax | NoneType | None | makes it possible to pass matplotlib axis to easily configure and save plots |
ax = plot_mutations(gen_list, ref_seq, sample_name="sacB", plot_range=[0,139], TR_range=[50,119])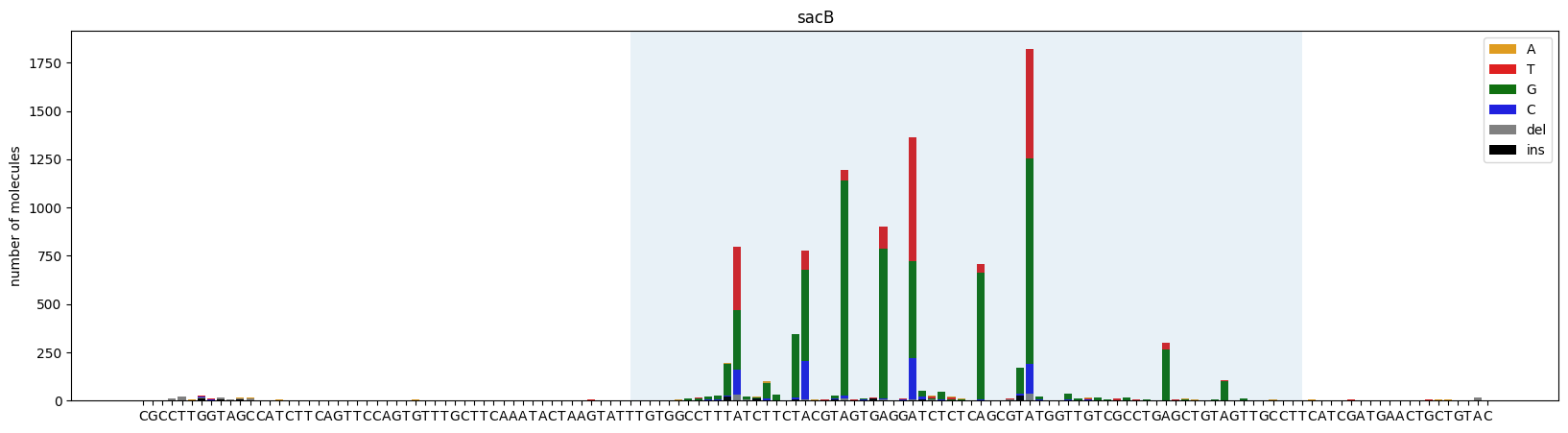
ax,mut_perc=plot_mutations_percentage(gen_list, ref_seq, sample_name="sacB", plot_range=[0,139], TR_range=[50,119])
plt.show
print(mut_perc)9.73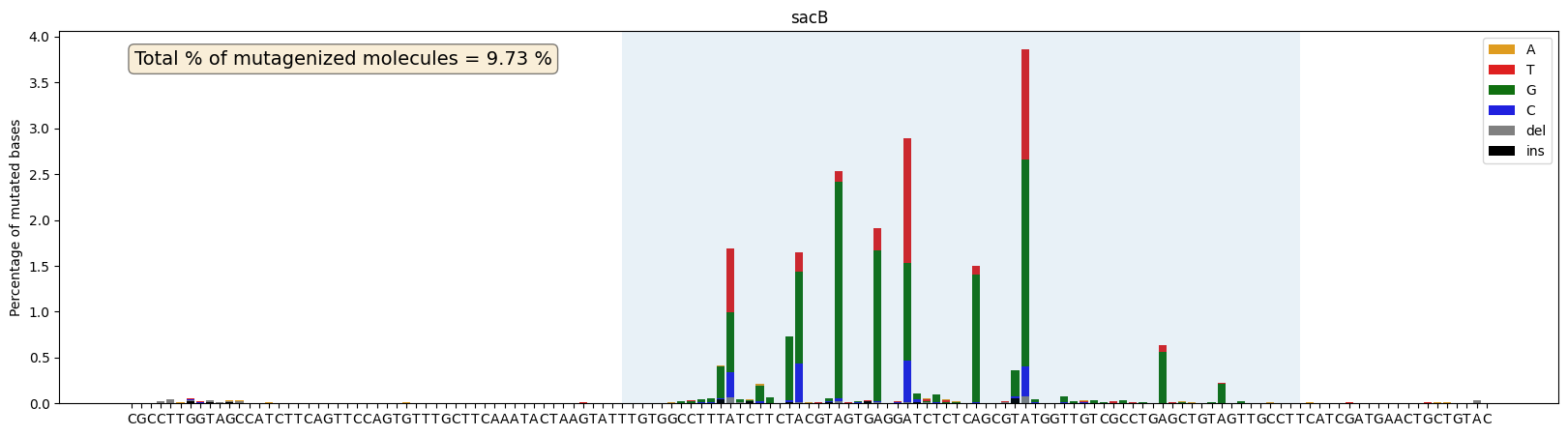
ax = plot_mutations_percentage(gen_list, ref_seq, sample_name="sacB", plot_range=[0,139], TR_range=[20,89],rev_comp=True)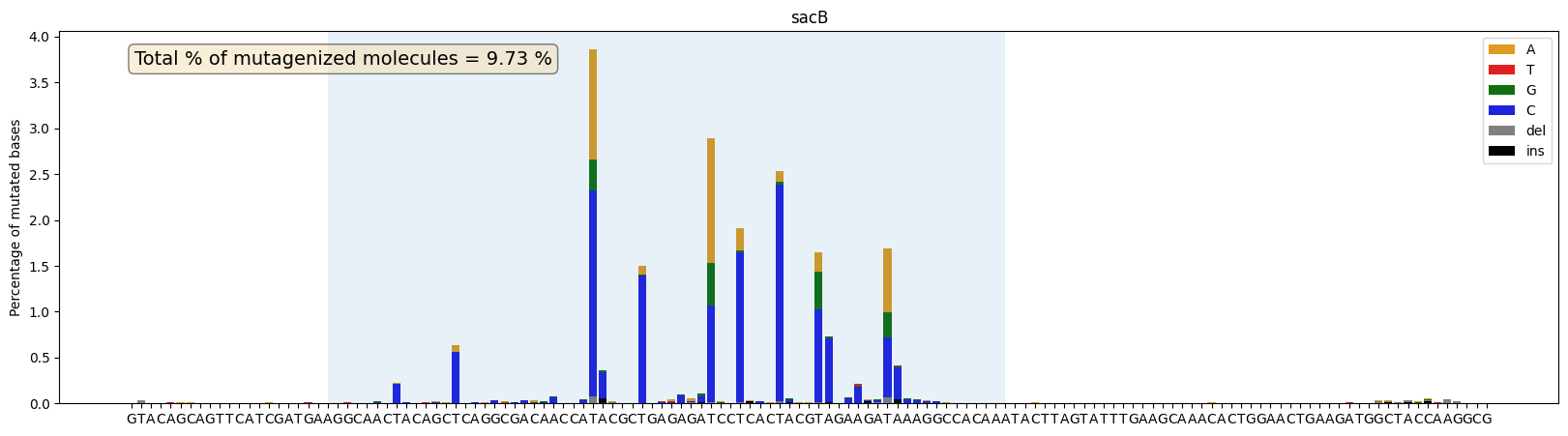
plot_mutations_percentage_protein
plot_mutations_percentage_protein (aa_mut_list, ref_prot, plot_range=None, ax=None)
| Type | Default | Details | |
|---|---|---|---|
| aa_mut_list | list of genotypes. Each genotype is a tuple: (string representation of the genotype, number of molecules) | ||
| ref_prot | reference sequence | ||
| plot_range | NoneType | None | limits the plot to the specified range |
| ax | NoneType | None |
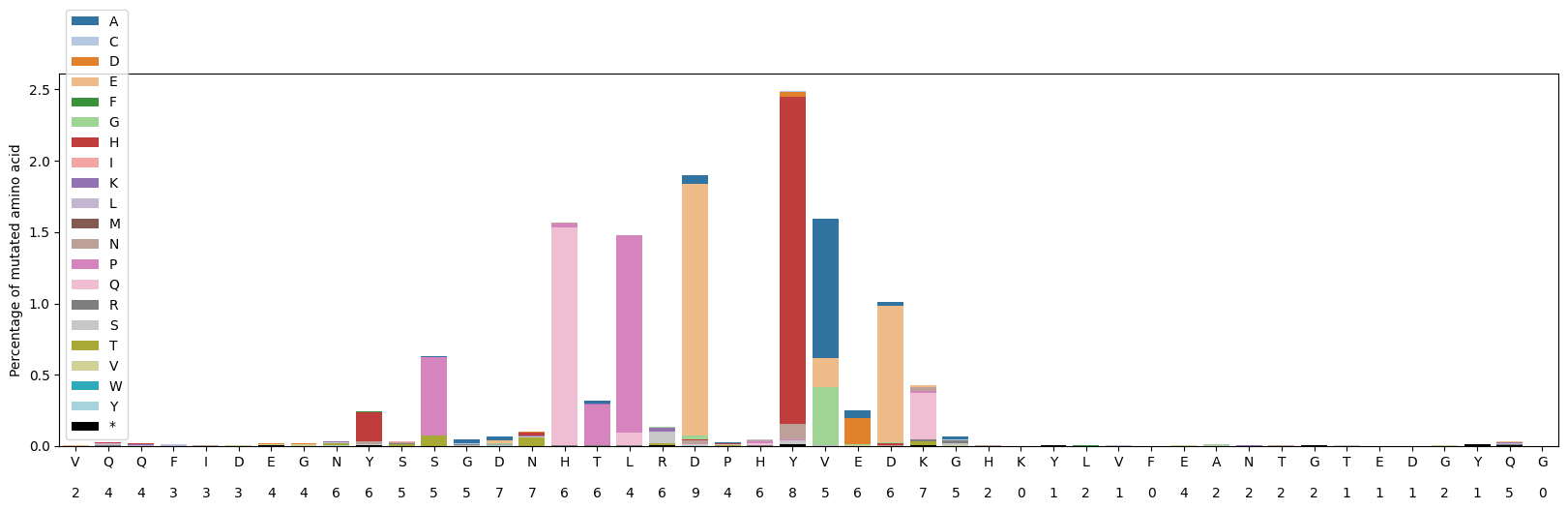
aa_mut_list=get_aa_mut_list(gen_list,ref_seq,ori=-1) #the sacB gene is in reverse complement orientation compared to the VR so ori=-1 is needed
aa_mut_list[:10][('', 43341),
('Y22H', 351),
('H15Q', 277),
('D19E', 246),
('L17P', 200),
('V23A', 162),
('S11P', 117),
('D25E', 113),
('D19E,Y22H', 75),
('T16P', 61)]ref_prot=Seq(ref_seq).reverse_complement()[:-1].translate() #the sacB gene is in reverse complement orientation compared to the VR
fig, ax = plt.subplots(1, 1, figsize=(20, 5))
plot_mutations_percentage_protein(aa_mut_list, ref_prot, ax=ax)
plt.show()/tmp/ipykernel_1816/3434536392.py:11: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap(obj)`` instead.
colormap = cm.get_cmap(colormap_name, num_colors)